Ultra-high resolution X-ray crystallography
Atomic resolution (<1.2 Å) X-ray crystallography reveals a wealth of structural details such as alternative residue conformations, anisotropic temperature factors, and hydrogen atom positions. This information allows us to study local protein movement and side chain protonation states essential for enzyme catalysis and ligand binding. Currently the highest resolution structure solved in our laboratory is 0.79 Å using CTX-M β-lactamase crystals. Our ultimate goal is to push the resolution to subatomic level (<0.7 Å) and, in combination with neutron crystallography, to elucidate macromolecular interactions and reactions at the level of charge density distribution.
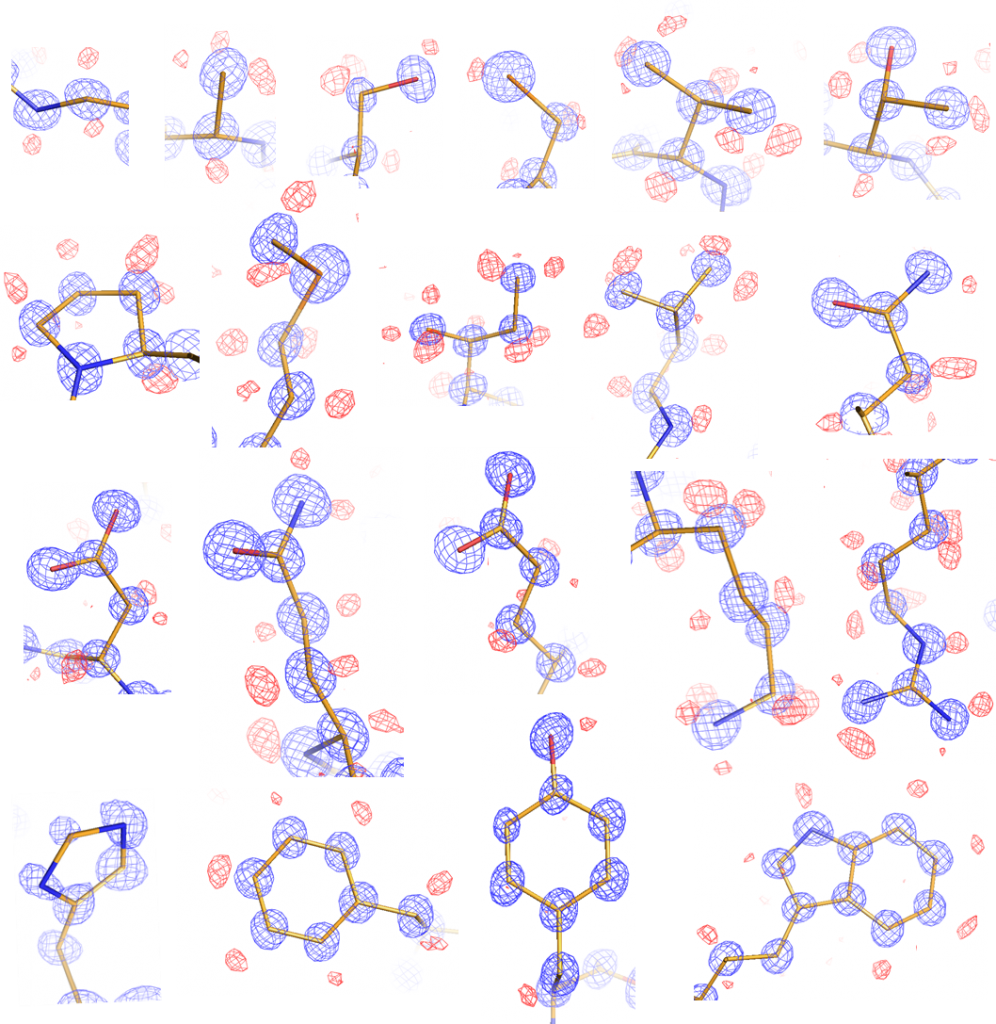 Figure 1. Ultra-high resolution of the common 20 amino acid side chains.
Figure 1. Ultra-high resolution of the common 20 amino acid side chains.
Fragment-based molecular docking
Screening for inhibitors using large drug like molecules (~500 Dalton MW) is inefficient as the number of theoretically possible compounds is > 10^60. Using a fragment-based approach offers a more efficient method to sample the vast amount of chemical space by first identifying low MW ligands and then linking or growing them into more specific, higher-affinity inhibitors. While traditional fragment screening is often low through-put due to experimental and financial restraints, we are interested in developing and applying computational docking in prioritizing fragments for experimental testing and in subsequent lead optimization. Combined with X-ray crystallography, this platform has allowed us to develop novel inhibitors against many challenging targets including bacterial proteins and protein-protein interfaces.
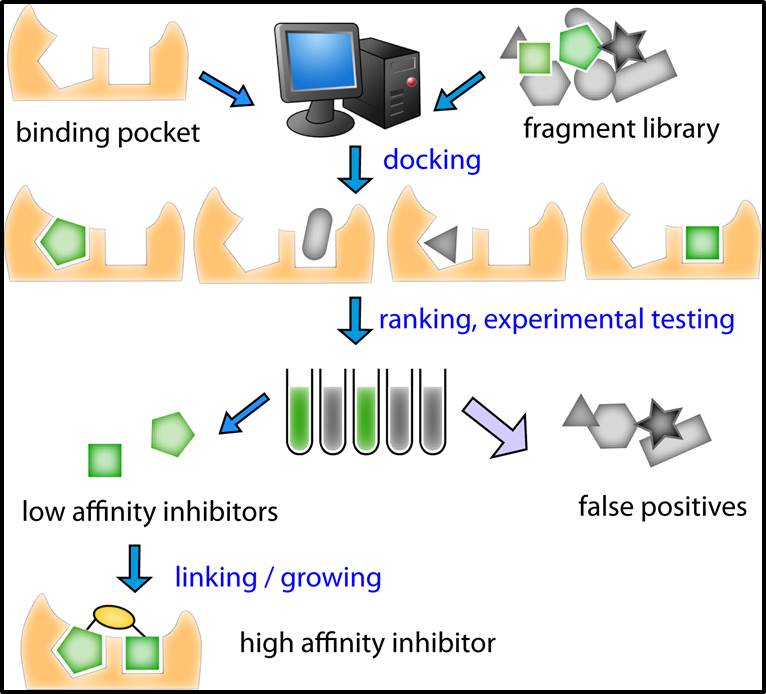 Figure 2. Scheme used to dock, rank, test, and optimize fragments for inhibition.
Figure 2. Scheme used to dock, rank, test, and optimize fragments for inhibition.
Low-barrier hydrogen bonds in protein stability and catalysis
Low barrier hydrogen bonds (LBHB) are a special type of short (~2.5 Å in length) and strong hydrogen bond (HB) that is characterized by the two electronegative atoms’ equal sharing of a proton. Because its strength can be 2-5 times greater than that of a normal HB, LBHB’s have been proposed to lower the energy barrier of enzymatic reactions. Our discovery of two potential LBHB’s in apo and complex CTX-M structures represent the first use of X-ray crystallography to directly identify hydrogen atoms engaged in such HB’s in proteins. These two instances include one in the CTX-M active site, between the catalytic Ser70 and Lys73, and another one between Asp233 and Asp246, which are two buried charged residues outside the active site. We are investigating the electrostatic and steric origins of these LBHB’s, and how they contribute to protein structure and function.
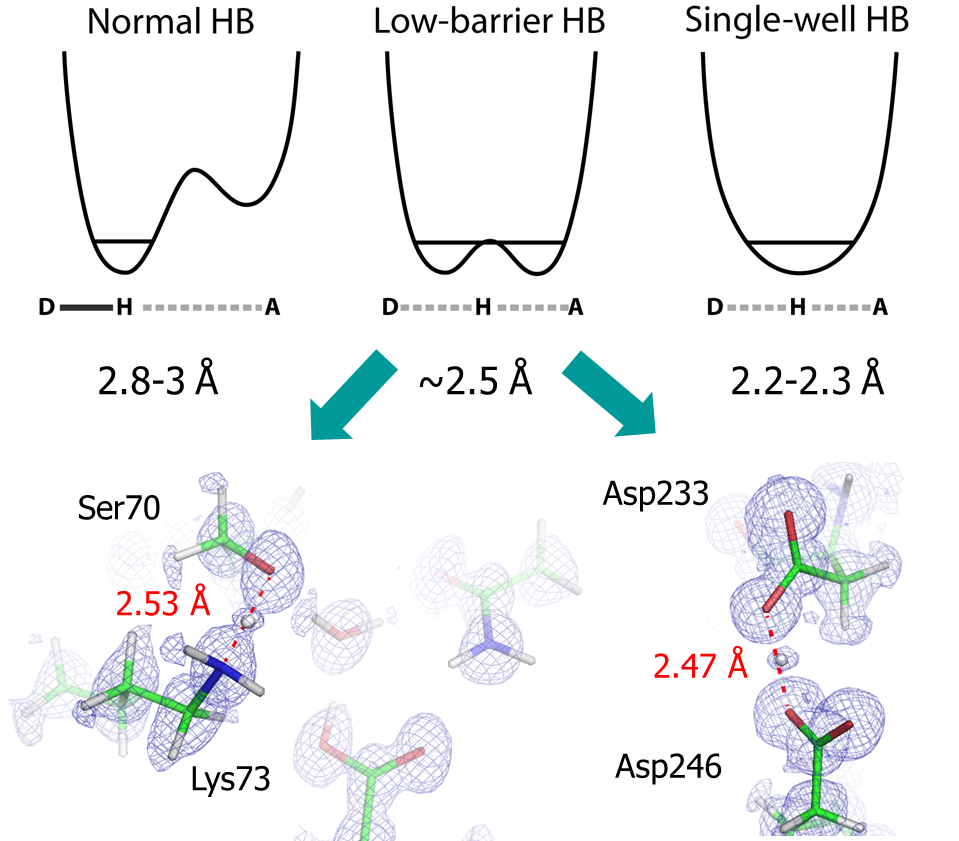 Figure 3. Low-barrier hydrogen bond between Ser70/Lys73 and Asp233/Asp246 of CTX-M beta-lactamase.
Figure 3. Low-barrier hydrogen bond between Ser70/Lys73 and Asp233/Asp246 of CTX-M beta-lactamase.
Novel antibiotic development targeting bacterial cell wall synthesis
The polymerization of bacterial cell wall peptidoglycan (a NAG-NAM disaccharide bearing a pentapeptide) consists of a transglycosylation reaction and followed by a transpeptidation reaction, the latter of which is inhibited by beta-lactam antibiotics. The beta-lactam antibiotics are the most-widely used antibiotics, but bacterial resistance to them is increasing, thanks in large part to bacterial production of beta-lactamases. In order to combat the increasingly urgent problem of antibiotic resistance, we are focused on developing novel non-covalent inhibitors against beta-lactamases, as well as other enzymes involved in bacterial cell wall synthesis, such as the penicillin binding proteins (PBPs).
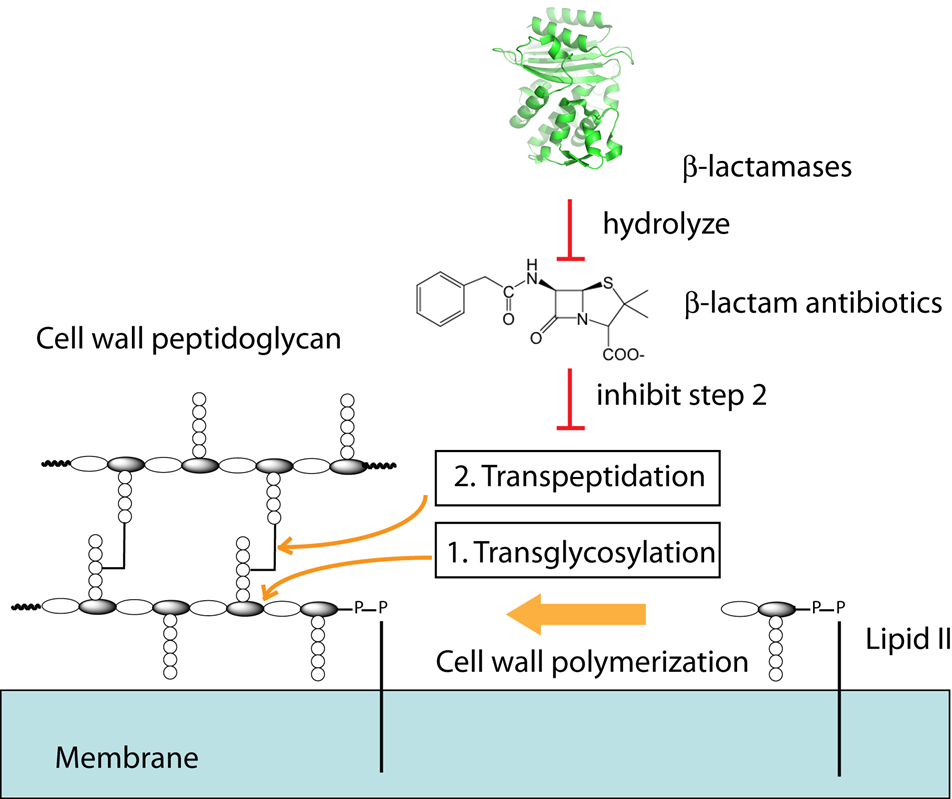
Figure 4. Bacterial cell wall synthesis is inhibited by beta-lactam antibiotics such as penicillin and offers a viable target for future antibiotic development.
Translational research: Cancer, Alzheimer’s disease, etc…
In collaboration with our colleagues at USF and other institutions, we have used our multidisciplinary platform to begin developing novel inhibitors against proteins involved in a range of human diseases. For example, we have uncovered small molecule ligands against CXCL12, which is a human chemokine protein that binds to CXCR4 GPCR and is responsible for the migration of tumor cells in metastatic cancer. Our inhibitors are able to disrupt the interactions between CXCL12 and CXCR4, and may provide new avenues for not only treating tumor metastasis, but studying cell migration in such diseases as well. Lead compounds have also been identified and are currently under development for proteins involved in Alzheimer’s and other diseases.
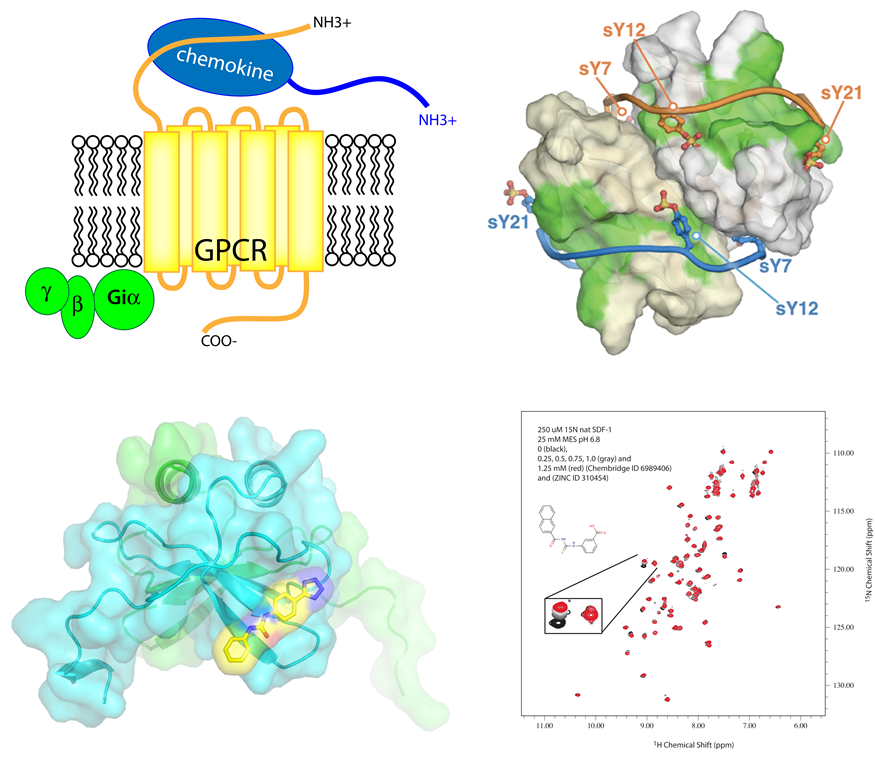 Figure 5. Development of novel small molecule inhibitors against human chemokine CXCL12.
Figure 5. Development of novel small molecule inhibitors against human chemokine CXCL12.
